CMIP6#
This notebook demonstrates access to Coupled Model Intercomparison Project Phase 6 (CMIP6) data. Broad information about the dataset can be found on the PACE website (see here)
Requirements to run this notebook
None
Objectives
Use pydap’s client API to demonstrate
To demonstrate remote access to CMIP data available through the **Earth System Grid Federation ESGF Portal.
To access and subset remote data using the DAP2 Protocol.
The Earth System Grid Federation ESGF Contains a broad range of model output (e.g, CMIP3, CMIP5, CMIP6, E3SM) from which you can obtain OPeNDAP URLs for data variables. To access the ESGF Node and browse data click here.
Author: Miguel Jimenez-Urias, ‘24
import matplotlib.pyplot as plt
import numpy as np
from pydap.client import open_url
import cartopy.crs as ccrs
CMIP6 Access via OPeNDAP server
You can also directly inspect a THREDDS catalog for CMIP6. For example, you can navigate to CDRMIP/CCCma/CanESM5/esm-pi-cdr-pulse/r2i1p2f1/Eday/ts/gn/v20190429 and access ts data via OPeNDAP DAP2 protocol.
url = "https://crd-esgf-drc.ec.gc.ca/thredds/dodsC/esgB_dataroot/AR6/CMIP6/CDRMIP/CCCma/CanESM5/esm-pi-cdr-pulse/r2i1p2f1/Eday/ts/gn/v20190429/ts_Eday_CanESM5_esm-pi-cdr-pulse_r2i1p2f1_gn_54510101-56501231.nc"
Create dataset access via pydap
By default protocol='dap2', however the default behavior may change in the future.
%%time
ds = open_url(url, protocol='dap2', output_grid=True)
CPU times: user 45.6 ms, sys: 9.27 ms, total: 54.8 ms
Wall time: 605 ms
ds.tree()
.esgB_dataroot/AR6/CMIP6/CDRMIP/CCCma/CanESM5/esm-pi-cdr-pulse/r2i1p2f1/Eday/ts/gn/v20190429/ts_Eday_CanESM5_esm-pi-cdr-pulse_r2i1p2f1_gn_54510101-56501231.nc
├──time
├──time_bnds
├──lat
├──lat_bnds
├──lon
├──lon_bnds
└──ts
├──ts
├──time
├──lat
└──lon
print('Dataset memory user [GBs, uncompressed]: ', ds.nbytes/1e9)
Dataset memory user [GBs, uncompressed]: 2.394406144
Inspect single variable
ts = ds['ts']
ts
<GridType with array 'ts' and maps 'time', 'lat', 'lon'>
Grid Arrays
No longer implemented in
DAP4. These carry copies of dimensions/coverage, and can be considered self-contained.Attempting to download into memory
tsalso downloadstime,lat,lon.Attributes sit the
GridTypelevel. For example:
ds['ts'].attributes
and
ds['ts']['ts'].attributes
yield different results.
def decode(variable) -> np.ndarray:
"""Decodes the variable BaseType according with atributes:
_FillValue
scale_factor
Parameters:
variable: BaseType (pydap model)
"""
import pydap
scale_factor = 1
_Fillvalue = None
if 'scale_factor' in variable.attributes:
scale_factor = variable.scale_factor
if '_FillValue' in variable.attributes:
if isinstance(variable, pydap.model.GridType):
data = np.where(variable.array.data == variable._FillValue, np.nan, variable.array.data)
elif isinstance(variable, pydap.model.BaseType):
data = np.where(variable.data == variable._FillValue, np.nan, variable.data)
else:
data = variable.data
return scale_factor * data
ts.tree()
.ts
├──ts
├──time
├──lat
└──lon
Let’s make some plots!
We will pick the Grid type ts at time=0. Will use pydap.
NOTE: When making a plot, check for missing values, scale factors, units.
ds['ts'].shape
(73000, 64, 128)
# download the entire GridType, single snapshot
GTS = ds['ts'][0, :, :]
GTS
<GridType with array 'ts' and maps 'time', 'lat', 'lon'>
GTS.attributes
{'standard_name': 'surface_temperature',
'long_name': 'Surface Temperature',
'comment': 'Temperature of the lower boundary of the atmosphere',
'units': 'K',
'original_name': 'GT',
'cell_methods': 'area: time: mean',
'cell_measures': 'area: areacella',
'history': '2019-08-20T21:03:55Z altered by CMOR: Reordered dimensions, original order: lat lon time. 2019-08-20T21:03:55Z altered by CMOR: replaced missing value flag (1e+38) and corresponding data with standard missing value (1e+20).',
'missing_value': 1e+20,
'_FillValue': 1e+20,
'_ChunkSizes': [1, 64, 128]}
GTS.shape
(1, 64, 128)
len(GTS.data), type(GTS.data)
(4, list)
NOTE:
# why is it greater than 1?
len(GTS.data) != 1
this is because downloading the Grid array downloads its coordinate dimensions too, resulting in a list of length 4!
New Default behavior#
Note
As of version 3.5.4, the default behavior now is output_grid=False, when using open_url . This has a very different behavior as with output_grid=True
%%time
ds = open_url(url, protocol='dap2') # output_grid=False as defaiult
CPU times: user 20 ms, sys: 3.67 ms, total: 23.6 ms
Wall time: 530 ms
# download the entire GridType, single snapshot
TS = ds['ts'][0, :, :]
TS
<BaseType with data array(shape=(1, 64, 128), dtype=>f4)>
len(TS.data), type(TS.data)
(1, numpy.ndarray)
NOTE:
len(TS.data) == 1
And TS is a BaseType, this means it did not download coordinate data when slicing the array
Note
Since the data is periodic in longitude, we need to append a copy to the array. We need to do this since cartopy interpolates data. If we don’t, then there will be missing longitude band of missing data in for plot as shown below:
Lon, Lat = np.meshgrid(ds['lon'].data, ds['lat'].data)
plt.figure(figsize=(15, 5))
ax = plt.axes(projection=ccrs.Mollweide())
ax.set_global()
ax.coastlines()
ax.contourf(Lon, Lat, np.squeeze(decode(GTS)), 200, transform=ccrs.PlateCarree(), cmap='jet')
plt.show()
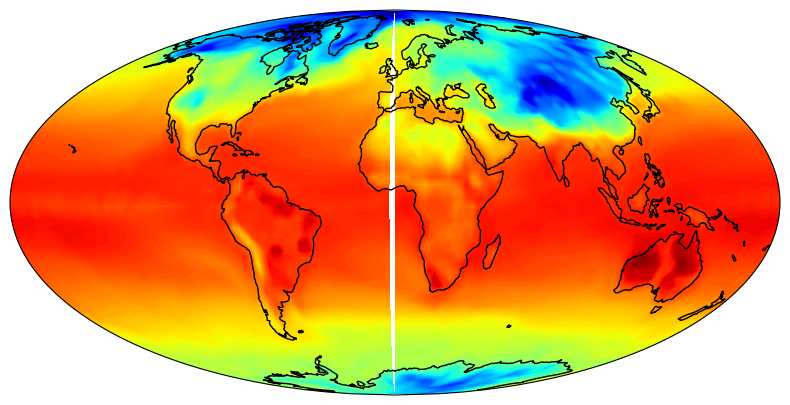
Fig 1. Global Near surface temperature on a (longitude)-periodic domain.
