Acceso a ECCOv4 por medio de EarthData in the Cloud de la NASA#
Este tutorial demuestra acceso al producto ECCOv4 de un modelo numerico global. Para mayor informacion del producto de ECCO, puede ir aqui.
Requisitos
Tener una cuenta de Earth Data Login por medio de la NASA.
Tener un Token valido.
Tambien se puede utilizar el metodo de Nombre de Usuario / Contrasena descrito en el tutorial de Autenticacion
Objectivos
Utilizar pydap para demostrar
Acceso to archivos cientificos de la NASA por medio del uso de
tokensy EarthData como metodo de autenticacion.
Algunas variables de interes son:
Autor: Miguel Jimenez-Urias, ‘24
import matplotlib.pyplot as plt
import numpy as np
from pydap.net import create_session
from pydap.client import open_url
import xarray as xr
import pydap
print("xarray version: ", xr.__version__)
print("pydap version: ", pydap.__version__)
xarray version: 2025.9.0
pydap version: 3.5.8
Acceso a EARTHDATA#
El listado de las variables de este producto pueden encontrarse aqui. En este caso, accederemos a las variables en su malla original, el LL90.
Grid_url = 'https://opendap.earthdata.nasa.gov/providers/POCLOUD/collections/ECCO%20Geometry%20Parameters%20for%20the%20Lat-Lon-Cap%2090%20(llc90)%20Native%20Model%20Grid%20(Version%204%20Release%204)/granules/GRID_GEOMETRY_ECCO_V4r4_native_llc0090'
Autenticacion via .netrc#
Las credenciales son recuperadas automaticamente por pydap.
my_session = create_session()
Alternativa: El use the tokens#
session_extra = {"token": "YourToken"}
# initialize a requests.session object with the token headers. All handled by pydap.
my_session = create_session(session_kwargs=session_extra)
Accesso a los metadatos solamente, por medio de pydap#
pydap aprovecha el protocolo de OPeNDAP, el cual permite la separacion de los metadatos de los valores numericos. Esto permite una inspeccion remota de los datasets.
ds_grid = open_url(Grid_url, session=my_session, protocol="dap4")
ds_grid.tree()
.GRID_GEOMETRY_ECCO_V4r4_native_llc0090.nc
├──dxC
├──PHrefF
├──XG
├──dyG
├──rA
├──hFacS
├──Zp1
├──Zl
├──rAw
├──dxG
├──maskW
├──YC
├──XC
├──maskS
├──YG
├──hFacC
├──drC
├──drF
├──XC_bnds
├──Zu
├──Z_bnds
├──YC_bnds
├──PHrefC
├──rAs
├──Depth
├──dyC
├──SN
├──rAz
├──maskC
├──CS
├──hFacW
├──Z
├──i
├──i_g
├──j
├──j_g
├──k
├──k_l
├──k_p1
├──k_u
├──nb
├──nv
└──tile
Note
PyDAP accesa solo a los metadatos de archivo en el servidor de OPeNDAP, y ningun arreglo numerico ha sido descargardo hasta este punto!
Para descargar los arreglos numericos hay que indexar la variable
Esto es, de la siguiente manera:
# this fetches remote data into a pydap object container
pydap_Array = dataset[<VarName>][:]
donde <VarName> es el nombre de una de las variables. El producto sera una representation “en memoria” de tipo pydap.model.BaseType, el cual permite el acceso a los arreglos de numpy.
Para extraer los valores de la variable remota, hay que ejecutar el siguiente comando
# The `.data` command allows direct access to the Numpy array (e.g. for manipulation)
pydap_Array.data
# lets download some data
Depth = ds_grid['Depth'][:]
print(type(Depth))
<class 'pydap.model.BaseType'>
Depth.attributes
{'_FillValue': 9.969209968e+36,
'long_name': 'model seafloor depth below ocean surface at rest',
'units': 'm',
'coordinate': 'XC YC',
'coverage_content_type': 'modelResult',
'standard_name': 'sea_floor_depth_below_geoid',
'comment': "Model sea surface height (SSH) of 0m corresponds to an ocean surface at rest relative to the geoid. Depth corresponds to seafloor depth below geoid. Note: the MITgcm used by ECCO V4r4 implements 'partial cells' so the actual model seafloor depth may differ from the seafloor depth provided by the input bathymetry file.",
'coordinates': 'YC XC',
'origname': 'Depth',
'fullnamepath': '/Depth',
'Maps': (),
'_DAP4_Checksum_CRC32': np.uint32(3811813944)}
Depth.shape, Depth.dims
((13, 90, 90), ['/tile', '/j', '/i'])
Visualizando el fondo oceanico Depth en la malla original del modelo
En este caso, el producto ECCO esta definido en una malla con topologia de un Cubo Esferico. De esta manera, la malla horizontal contiene una dimension extra llamada: tile o face. A continuacion visualizamos la variable en su topologia original
Variable = [Depth[i].data for i in range(13)]
clevels = np.linspace(0, 6000, 100)
cMap = 'Greys_r'
fig, axes = plt.subplots(nrows=5, ncols=5, figsize=(8, 8), gridspec_kw={'hspace':0.01, 'wspace':0.01})
AXES = [
axes[4, 0], axes[3, 0], axes[2, 0], axes[4, 1], axes[3, 1], axes[2, 1],
axes[1, 1],
axes[1, 2], axes[1, 3], axes[1, 4], axes[0, 2], axes[0, 3], axes[0, 4],
]
for i in range(len(AXES)):
AXES[i].contourf(Variable[i], clevels, cmap=cMap)
for ax in np.ravel(axes):
ax.axis('off')
plt.setp(ax.get_xticklabels(), visible=False)
plt.setp(ax.get_yticklabels(), visible=False)
plt.show()
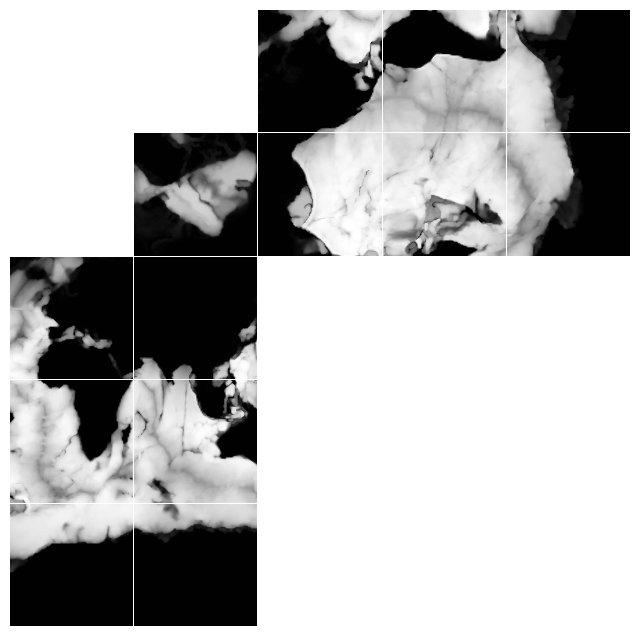
Fig. 1. La variable Depth visualizada en una malla horitonzal. Tiles con valores 0-5 tienen un ordenamiento de valores (indiciales) C-ordering, mientras que cualquier arreglo numerico tiles 7-13 siguen el F-ordering. Cualquier arreglo numerico en el arctic cap, tiene un comportamiento de una coordenada polar.
Visualizacion con topologia corregida
fig, axes = plt.subplots(nrows=4, ncols=4, figsize=(8, 8), gridspec_kw={'hspace':0.01, 'wspace':0.01})
AXES_NR = [
axes[3, 0], axes[2, 0], axes[1, 0], axes[3, 1], axes[2, 1], axes[1, 1],
]
AXES_CAP = [axes[0, 0]]
AXES_R = [
axes[1, 2], axes[2, 2], axes[3, 2], axes[1, 3], axes[2, 3], axes[3, 3],
]
for i in range(len(AXES_NR)):
AXES_NR[i].contourf(Variable[i], clevels, cmap=cMap)
for i in range(len(AXES_CAP)):
AXES_CAP[i].contourf(Variable[6].transpose()[:, ::-1], clevels, cmap=cMap)
for i in range(len(AXES_R)):
AXES_R[i].contourf(Variable[7+i].transpose()[::-1, :], clevels, cmap=cMap)
for ax in np.ravel(axes):
ax.axis('off')
plt.setp(ax.get_xticklabels(), visible=False)
plt.setp(ax.get_yticklabels(), visible=False)
plt.show()
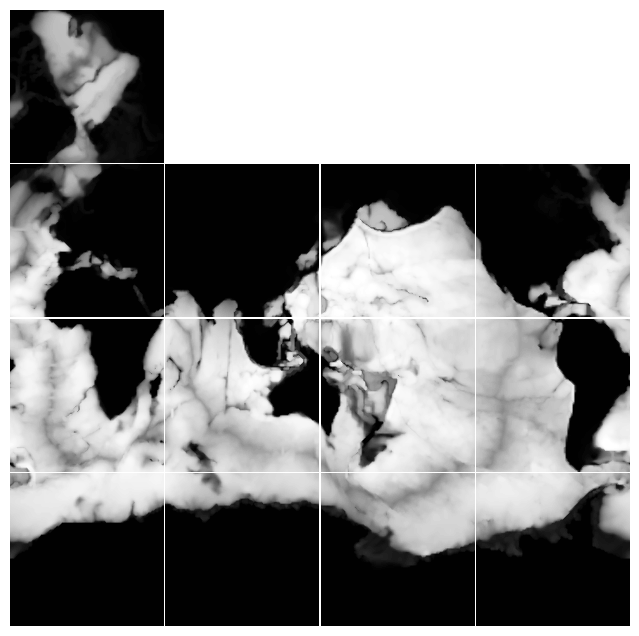
Fig. 2. Visualizacion de la variable Depth en una malla horizontal que approxima una malla horizontal uniforme con ejes lat-lon. Sin embargo, cualquier arreglo numerico en el arctic cap, continua en una malla que approxima una malla en coordenadas polares
Utilizando xarray#
pydap se puede llamar dentro de xarray para acceder a los archivos expuestos mediante el servidor de OPeNDAP. En particular, xarray permite aggregar multiples archivos remotos de opendap. A continuacion demostramos un pequeno ejemplo.
Un URL individual –> un archivo remoto
Below we access remote Temperature and Salinity ECCO data with xarray via (internally) pydap.
baseURL = 'https://opendap.earthdata.nasa.gov/providers/POCLOUD/collections/'
Temp_Salt = "ECCO%20Ocean%20Temperature%20and%20Salinity%20-%20Monthly%20Mean%20llc90%20Grid%20(Version%204%20Release%204)/granules/OCEAN_TEMPERATURE_SALINITY_mon_mean_"
year = '2017-'
month = '01'
end_ = '_ECCO_V4r4_native_llc0090'
Temp_2017 = baseURL + Temp_Salt +year + month + end_
# Convertimos el URL en un DAP4 opendap url
Temp_2017 = Temp_2017.replace("https", "dap4")
Temp_2017
'dap4://opendap.earthdata.nasa.gov/providers/POCLOUD/collections/ECCO%20Ocean%20Temperature%20and%20Salinity%20-%20Monthly%20Mean%20llc90%20Grid%20(Version%204%20Release%204)/granules/OCEAN_TEMPERATURE_SALINITY_mon_mean_2017-01_ECCO_V4r4_native_llc0090'
pyds = open_url(Temp_2017, session=my_session)
pyds.tree()
.OCEAN_TEMPERATURE_SALINITY_mon_mean_2017-01_ECCO_V4r4_native_llc0090.nc
├──XG
├──Zp1
├──Zl
├──YC
├──XC
├──SALT
├──YG
├──XC_bnds
├──Zu
├──THETA
├──Z_bnds
├──YC_bnds
├──time_bnds
├──Z
├──i
├──i_g
├──j
├──j_g
├──k
├──k_l
├──k_p1
├──k_u
├──nb
├──nv
├──tile
└──time
Para evitar descargas variables que no sean necesariamente de interest, se le puede instruir al Servidor Hyrax que variables requiere uno.
Para esto utilizaremos las Expresiones de Restriccion (CE, por sus siglas en ingles). Este metodo nos ayudara a construir datasets mucho mas simples,
y evitar descargar N veces, informacion que no es requerida.
A continuacion demostramos el caso de solo requerir la variable THETA y sus dimensiones.
dims = pyds['/THETA'].dims
Vars = ['/THETA'] + dims
# Below construct Contraint Expression
CE = "?dap4.ce="+(";").join(Vars)
print("Constraint Expression: ", CE)
Constraint Expression: ?dap4.ce=/THETA;/time;/k;/tile;/j;/i
Temp_2017 = baseURL + Temp_Salt +year + month + end_ + CE
Temp_2017
'https://opendap.earthdata.nasa.gov/providers/POCLOUD/collections/ECCO%20Ocean%20Temperature%20and%20Salinity%20-%20Monthly%20Mean%20llc90%20Grid%20(Version%204%20Release%204)/granules/OCEAN_TEMPERATURE_SALINITY_mon_mean_2017-01_ECCO_V4r4_native_llc0090?dap4.ce=/THETA;/time;/k;/tile;/j;/i'
Importante#
Ahora aplicamos la CE a todos los possibles URLs the nuestro dataset.
Temp_2017 = [baseURL.replace("https", "dap4") + Temp_Salt + year + f'{i:02}' + end_ + CE for i in range(1, 13)]
Temp_2017[:3]
['dap4://opendap.earthdata.nasa.gov/providers/POCLOUD/collections/ECCO%20Ocean%20Temperature%20and%20Salinity%20-%20Monthly%20Mean%20llc90%20Grid%20(Version%204%20Release%204)/granules/OCEAN_TEMPERATURE_SALINITY_mon_mean_2017-01_ECCO_V4r4_native_llc0090?dap4.ce=/THETA;/time;/k;/tile;/j;/i',
'dap4://opendap.earthdata.nasa.gov/providers/POCLOUD/collections/ECCO%20Ocean%20Temperature%20and%20Salinity%20-%20Monthly%20Mean%20llc90%20Grid%20(Version%204%20Release%204)/granules/OCEAN_TEMPERATURE_SALINITY_mon_mean_2017-02_ECCO_V4r4_native_llc0090?dap4.ce=/THETA;/time;/k;/tile;/j;/i',
'dap4://opendap.earthdata.nasa.gov/providers/POCLOUD/collections/ECCO%20Ocean%20Temperature%20and%20Salinity%20-%20Monthly%20Mean%20llc90%20Grid%20(Version%204%20Release%204)/granules/OCEAN_TEMPERATURE_SALINITY_mon_mean_2017-03_ECCO_V4r4_native_llc0090?dap4.ce=/THETA;/time;/k;/tile;/j;/i']
#
Debajo inicializaremos una session que persista, cada vez que descargue datos de servidores de OPeNDAP. En este caso utilizaremost la libraria
requests_cache. El comportamiento es casi identico excepto la siguiente celda:
cache_session = create_session(use_cache=True, cache_kwargs={"cache_name":"data/ecco"}) # caching the session
cache_session.cache.clear() # clear all cache to demonstrate the behavior
Warning
A partir de la version de pydap >= 3.5.5, existe el nuevo metodo experimental consolidated_metadata que permite a pydap descargar todos los metadatos, y reusarlos. Este metodo continua en desarrollo y sus propiedades pueden cambiar en las siguientes versiones.
from pydap.client import consolidate_metadata
%%time
consolidate_metadata(Temp_2017, session=cache_session, concat_dim='time')
datacube has dimensions ['i[0:1:89]', 'j[0:1:89]', 'k[0:1:49]', 'tile[0:1:12]'] , and concat dim: `['time']`
CPU times: user 1.11 s, sys: 401 ms, total: 1.51 s
Wall time: 18.4 s
%%time
theta_salt_ds = xr.open_mfdataset(
Temp_2017,
engine='pydap',
session=cache_session,
parallel=True,
combine='nested',
concat_dim='time',
)
CPU times: user 281 ms, sys: 149 ms, total: 430 ms
Wall time: 371 ms
---------------------------------------------------------------------------
ParseError Traceback (most recent call last)
[... skipping hidden 1 frame]
Cell In[21], line 1
----> 1 get_ipython().run_cell_magic('time', '', "theta_salt_ds = xr.open_mfdataset(\n Temp_2017, \n engine='pydap',\n session=cache_session, \n parallel=True, \n combine='nested', \n concat_dim='time',\n)\n")
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/interactiveshell.py:2565, in InteractiveShell.run_cell_magic(self, magic_name, line, cell)
2564 args = (magic_arg_s, cell)
-> 2565 result = fn(*args, **kwargs)
2567 # The code below prevents the output from being displayed
2568 # when using magics with decorator @output_can_be_silenced
2569 # when the last Python token in the expression is a ';'.
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/magics/execution.py:1452, in ExecutionMagics.time(self, line, cell, local_ns)
1451 if exit_on_interrupt and captured_exception:
-> 1452 raise captured_exception
1453 return
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/magics/execution.py:1416, in ExecutionMagics.time(self, line, cell, local_ns)
1415 try:
-> 1416 exec(code, glob, local_ns)
1417 out = None
File <timed exec>:1
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/xarray/backends/api.py:1812, in open_mfdataset(paths, chunks, concat_dim, compat, preprocess, engine, data_vars, coords, combine, parallel, join, attrs_file, combine_attrs, errors, **kwargs)
1809 if parallel:
1810 # calling compute here will return the datasets/file_objs lists,
1811 # the underlying datasets will still be stored as dask arrays
-> 1812 datasets, closers = dask.compute(datasets, closers)
1814 # Combine all datasets, closing them in case of a ValueError
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/dask/base.py:681, in compute(traverse, optimize_graph, scheduler, get, *args, **kwargs)
679 keys = list(flatten(expr.__dask_keys__()))
--> 681 results = schedule(expr, keys, **kwargs)
683 return repack(results)
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/xarray/backends/api.py:760, in open_dataset(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, create_default_indexes, inline_array, chunked_array_type, from_array_kwargs, backend_kwargs, **kwargs)
759 overwrite_encoded_chunks = kwargs.pop("overwrite_encoded_chunks", None)
--> 760 backend_ds = backend.open_dataset(
761 filename_or_obj,
762 drop_variables=drop_variables,
763 **decoders,
764 **kwargs,
765 )
766 ds = _dataset_from_backend_dataset(
767 backend_ds,
768 filename_or_obj,
(...) 779 **kwargs,
780 )
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/xarray/backends/pydap_.py:235, in PydapBackendEntrypoint.open_dataset(self, filename_or_obj, mask_and_scale, decode_times, concat_characters, decode_coords, drop_variables, use_cftime, decode_timedelta, group, application, session, output_grid, timeout, verify, user_charset)
214 def open_dataset(
215 self,
216 filename_or_obj: (
(...) 233 user_charset=None,
234 ) -> Dataset:
--> 235 store = PydapDataStore.open(
236 url=filename_or_obj,
237 group=group,
238 application=application,
239 session=session,
240 output_grid=output_grid,
241 timeout=timeout,
242 verify=verify,
243 user_charset=user_charset,
244 )
245 store_entrypoint = StoreBackendEntrypoint()
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/xarray/backends/pydap_.py:131, in PydapDataStore.open(cls, url, group, application, session, output_grid, timeout, verify, user_charset)
129 if isinstance(url, str):
130 # check uit begins with an acceptable scheme
--> 131 dataset = open_url(**kwargs)
132 elif hasattr(url, "ds"):
133 # pydap dataset
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/pydap/client.py:156, in open_url(url, application, session, output_grid, timeout, verify, checksums, user_charset, protocol, batch, use_cache, session_kwargs, cache_kwargs, get_kwargs)
150 session = create_session(
151 use_cache=use_cache,
152 session_kwargs=session_kwargs,
153 cache_kwargs=cache_kwargs,
154 )
--> 156 handler = DAPHandler(
157 url,
158 application,
159 session,
160 output_grid,
161 timeout=timeout,
162 verify=verify,
163 checksums=checksums,
164 user_charset=user_charset,
165 protocol=protocol,
166 get_kwargs=get_kwargs,
167 )
168 dataset = handler.dataset
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/pydap/handlers/dap.py:122, in DAPHandler.__init__(self, url, application, session, output_grid, timeout, verify, checksums, user_charset, protocol, get_kwargs)
121 self.base_url = urlunparse(arg)
--> 122 self.make_dataset()
123 self.add_proxies()
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/pydap/handlers/dap.py:157, in DAPHandler.make_dataset(self)
156 if self.protocol == "dap4":
--> 157 self.dataset_from_dap4()
158 else:
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/pydap/handlers/dap.py:186, in DAPHandler.dataset_from_dap4(self)
185 dmr = safe_charset_text(r, self.user_charset)
--> 186 self.dataset = dmr_to_dataset(dmr)
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/pydap/parsers/dmr.py:252, in dmr_to_dataset(dmr)
251 # Parse the DMR. First dropping the namespace
--> 252 dom_et = copy.deepcopy(DMRParser(dmr).node)
253 # emtpy dataset
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/pydap/parsers/dmr.py:357, in DMRParser.__init__(self, dmr)
356 _dmr = re.sub(' xmlns="[^"]+"', "", self.dmr, count=1)
--> 357 self.node = ET.fromstring(_dmr)
358 if len(get_groups(self.node)) > 0:
359 # no groups here
File ~/miniforge3/envs/pydap_docs/lib/python3.11/xml/etree/ElementTree.py:1350, in XML(text, parser)
1349 parser = XMLParser(target=TreeBuilder())
-> 1350 parser.feed(text)
1351 return parser.close()
ParseError: not well-formed (invalid token): line 1, column 0 (<string>)
During handling of the above exception, another exception occurred:
AssertionError Traceback (most recent call last)
[... skipping hidden 1 frame]
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/interactiveshell.py:2164, in InteractiveShell.showtraceback(self, exc_tuple, filename, tb_offset, exception_only, running_compiled_code)
2159 return
2161 if issubclass(etype, SyntaxError):
2162 # Though this won't be called by syntax errors in the input
2163 # line, there may be SyntaxError cases with imported code.
-> 2164 self.showsyntaxerror(filename, running_compiled_code)
2165 elif etype is UsageError:
2166 self.show_usage_error(value)
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/interactiveshell.py:2254, in InteractiveShell.showsyntaxerror(self, filename, running_compiled_code)
2252 # If the error occurred when executing compiled code, we should provide full stacktrace.
2253 elist = traceback.extract_tb(last_traceback) if running_compiled_code else []
-> 2254 stb = self.SyntaxTB.structured_traceback(etype, value, elist)
2255 self._showtraceback(etype, value, stb)
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/ultratb.py:1243, in SyntaxTB.structured_traceback(self, etype, evalue, etb, tb_offset, context)
1241 value.text = newtext
1242 self.last_syntax_error = value
-> 1243 return super(SyntaxTB, self).structured_traceback(
1244 etype, value, etb, tb_offset=tb_offset, context=context
1245 )
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/ultratb.py:212, in ListTB.structured_traceback(self, etype, evalue, etb, tb_offset, context)
210 out_list.extend(self._format_list(elist))
211 # The exception info should be a single entry in the list.
--> 212 lines = "".join(self._format_exception_only(etype, evalue))
213 out_list.append(lines)
215 # Find chained exceptions if we have a traceback (not for exception-only mode)
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/ultratb.py:341, in ListTB._format_exception_only(self, etype, value)
327 output_list.append(
328 theme_table[self._theme_name].format(
329 [(Token, " ")]
(...) 336 )
337 )
338 if textline == "":
339 # sep 2025:
340 # textline = py3compat.cast_unicode(value.text, "utf-8")
--> 341 assert isinstance(value.text, str)
342 textline = value.text
344 if textline is not None:
AssertionError:
---------------------------------------------------------------------------
AssertionError Traceback (most recent call last)
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/async_helpers.py:128, in _pseudo_sync_runner(coro)
120 """
121 A runner that does not really allow async execution, and just advance the coroutine.
122
(...) 125 Credit to Nathaniel Smith
126 """
127 try:
--> 128 coro.send(None)
129 except StopIteration as exc:
130 return exc.value
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/interactiveshell.py:3413, in InteractiveShell.run_cell_async(self, raw_cell, store_history, silent, shell_futures, transformed_cell, preprocessing_exc_tuple, cell_id)
3409 exec_count = self.execution_count
3410 if result.error_in_exec:
3411 # Store formatted traceback and error details
3412 self.history_manager.exceptions[exec_count] = (
-> 3413 self._format_exception_for_storage(result.error_in_exec)
3414 )
3416 # Each cell is a *single* input, regardless of how many lines it has
3417 self.execution_count += 1
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/interactiveshell.py:3442, in InteractiveShell._format_exception_for_storage(self, exception, filename, running_compiled_code)
3440 # Extract traceback if the error happened during compiled code execution
3441 elist = traceback.extract_tb(tb) if running_compiled_code else []
-> 3442 stb = self.SyntaxTB.structured_traceback(etype, evalue, elist)
3444 # Handle UsageError with a simple message
3445 elif etype is UsageError:
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/ultratb.py:1243, in SyntaxTB.structured_traceback(self, etype, evalue, etb, tb_offset, context)
1241 value.text = newtext
1242 self.last_syntax_error = value
-> 1243 return super(SyntaxTB, self).structured_traceback(
1244 etype, value, etb, tb_offset=tb_offset, context=context
1245 )
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/ultratb.py:212, in ListTB.structured_traceback(self, etype, evalue, etb, tb_offset, context)
210 out_list.extend(self._format_list(elist))
211 # The exception info should be a single entry in the list.
--> 212 lines = "".join(self._format_exception_only(etype, evalue))
213 out_list.append(lines)
215 # Find chained exceptions if we have a traceback (not for exception-only mode)
File ~/miniforge3/envs/pydap_docs/lib/python3.11/site-packages/IPython/core/ultratb.py:341, in ListTB._format_exception_only(self, etype, value)
327 output_list.append(
328 theme_table[self._theme_name].format(
329 [(Token, " ")]
(...) 336 )
337 )
338 if textline == "":
339 # sep 2025:
340 # textline = py3compat.cast_unicode(value.text, "utf-8")
--> 341 assert isinstance(value.text, str)
342 textline = value.text
344 if textline is not None:
AssertionError:
theta_salt_ds
Finalmente, visualizamos THETA#
Variable = [theta_salt_ds['THETA'][0, 0, i, :, :] for i in range(13)]
clevels = np.linspace(-5, 30, 100)
cMap='RdBu_r'
fig, axes = plt.subplots(nrows=4, ncols=4, figsize=(8, 8), gridspec_kw={'hspace':0.01, 'wspace':0.01})
AXES_NR = [
axes[3, 0], axes[2, 0], axes[1, 0], axes[3, 1], axes[2, 1], axes[1, 1],
]
AXES_CAP = [axes[0, 0]]
AXES_R = [
axes[1, 2], axes[2, 2], axes[3, 2], axes[1, 3], axes[2, 3], axes[3, 3],
]
for i in range(len(AXES_NR)):
AXES_NR[i].contourf(Variable[i], clevels, cmap=cMap)
for i in range(len(AXES_CAP)):
AXES_CAP[i].contourf(Variable[6].transpose()[:, ::-1], clevels, cmap=cMap)
for i in range(len(AXES_R)):
AXES_R[i].contourf(Variable[7+i].transpose()[::-1, :], clevels, cmap=cMap)
for ax in np.ravel(axes):
ax.axis('off')
plt.setp(ax.get_xticklabels(), visible=False)
plt.setp(ax.get_yticklabels(), visible=False)
plt.show()
Fig. 3. Visualizacion de la variable Surface temperature, similar al metodo utilizado en la Figura 2.
